王玮
| 王玮,博士,教授 中国海洋大学方宗熙中心 PI 邮箱:wangweisfc@ouc.edu.cn
|
教育经历
1998-2002,安徽中医药大学药学院,药学,理学学士
2003-2006,复旦大学生命科学学院, 植物学,理学硕士
2006-2010,香港中文大学,生物学,博士
工作经历
2010-2021,纽约大学,博士后研究员 / 研究科学家
2021至今,中国海洋大学,方宗熙海洋生物进化与发育研究中心,PI
主要研究方向及成果
王玮课题组致力于从全转录组的尺度上诠释基因调控网络在器官构建过程中的调控机制。生物体器官的正常构建有赖于胚胎发育过程中相关调控基因表达在时空和空间尺度上的精确调控。据统计, 每年约有1%的新生儿会出现先天性心脏发育异常 (congenital heart defect,CHD)。本课题组以海洋模式动物海鞘为模型,综合运用细胞分离纯化,高通量单细胞测序,细胞谱系追踪以及生物信息学分析等技术手段,力求系统性阐明心脏发育过程中的分子调控机制,为预防以及治疗先天性心脏发育异常提供理论支撑。王玮教授为主要研究人员承担多项由NIH/NHLBI,NIH/NICHD 以及Leducq Foundation – Trans-Atlantic Network of Excellence 资助的重大研究攻关项目,研究成果发表于Nature Cell Biology,PLOS Biology,Developmental Cell,Nature Communication,eLife,Developmenal Biology, PLOS Computational Biology,Current Topics in Developmental Biology, Advances in Experimental Medicine and Biology,Frontiers in Cell and Developmental Biology等期刊。
王玮教授应邀担任Developmental Biology, Developmental Dynamics, BMC Genomics, JEZ Part B: Molecular and Developmental Evolution 等期刊的审稿人,并于2018年被Developmental Biology 授予 “Outstanding Contribution in Reviewing”奖项。王玮教授应邀于2020担任National Science Foundation (NSF) Graduate Research Fellowship Program (GRFP) 评审人。王玮教授于2021年担任 Frontiers in Cell and Developmental Biology客座编辑, 主持 “Toward a System-Level Understanding of Invertebrate Chordate Development”特刊编写。
单细胞测序勾画心脏发育调控机制
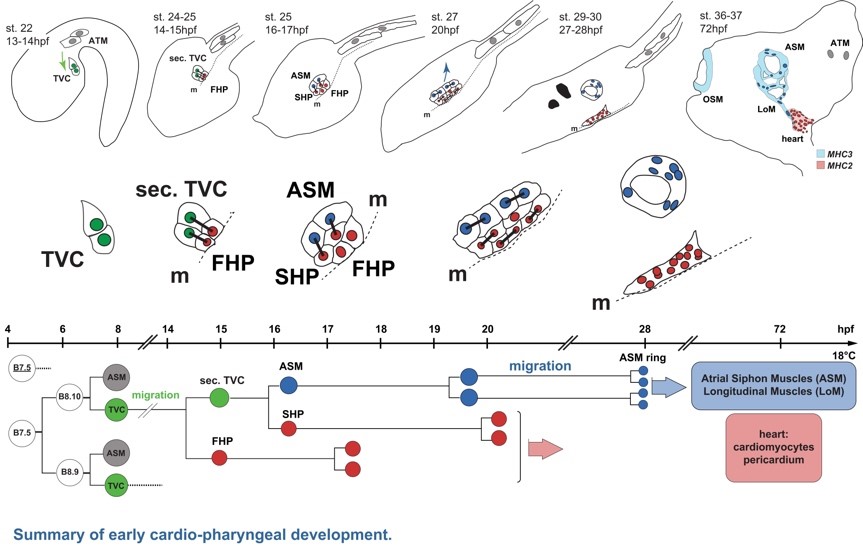
在脊椎动物心脏发育过程中,心肌和头部鳃节肌(branchiomeric head muscles)共同起源于位于咽中胚层(pharyngeal mesoderm)中的多能前体细胞群(multipotent progenitors)。但是由多能前体细胞分化为心脏中的多种功能细胞的转录调控机制仍有待系统性解析。
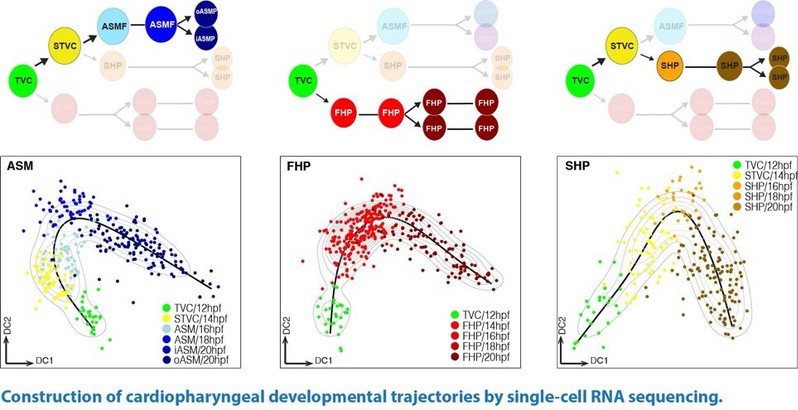
王玮教授应用单细胞测序技术,以海鞘作为模式动物,分别构建了心肌(first and second heart lineages)和咽肌(pharyngeal muscle)前体的分化轨迹(developmental trajectories),并从转录组尺度揭示了这一早期分化过程中的调控机制。
在这一研究的基础上,王玮课题组将进一步探讨心脏发育过程中的几个关键问题,包括:1)The First Heart Field (FHF) & Second Heart Field (SHF) 中前体细胞的多样性。2)各类前体细胞与成熟心脏中多种功能性细胞之间的发育关联。3)动态转录组以及染色质结构(chromatin architecture)在前体细胞命运决定中的调控机制。
课题组诚邀优秀科研人员加盟。
代表性文章
1.Nomaru H, Liu Y, De Bono C, Righelli D, Cirino A, Wang W, et al. Single cell multi-omic analysis identifies a Tbx1-dependent multilineage primed population in murine cardiopharyngeal mesoderm. Nature Communications, 2021;12: 6645.
2. Kim K, Gibboney S, Razy-Krajka F, Lowe EK, Wang W, Stolfi A. Regulation of neurogenesis by FGF signaling and neurogenin in the invertebrate chordate Ciona. Frontiers in Cell and Developmental Biology, 2020;8: 477.
3. Wang W, Niu X, Stuart T, Jullian E, Mauck WM 3rd, Kelly RG, et al. A single-cell transcriptional roadmap for cardiopharyngeal fate diversification. Nature Cell Biology, 2019;21: 674–686.
4. Kaplan NA, Wang W, Christiaen L. Initial characterization of Wnt-Tcf functions during Cionaheart development. Developmental Biology, 2019;448: 199–209.
5. Bernadskaya YY, Brahmbhatt S, Gline SE, Wang W, Christiaen L. Discoidin-domain receptor coordinates cell-matrix adhesion and collective polarity in migratory cardiopharyngeal progenitors. Nature Communications, 2019;10: 57.
6.Athanasiadou R, Neymotin B, Brandt N, Wang W, Christiaen L, Gresham D, et al. A complete statistical model for calibration of RNA-seq counts using external spike-ins and maximum likelihood theory. PLoS Computational Biology, 2019;15: e1006794.
7. Sharma S, Wang W, Stolfi A. Single-cell transcriptome profiling of theCionalarval brain. Developmental Biology, 2019;448: 226–236.
8. Razy-Krajka F, Gravez B, Kaplan N, Racioppi C, Wang W, Christiaen L. An FGF-driven feed-forward circuit patterns the cardiopharyngeal mesoderm in space and time. eLife. 2018;7. doi:10.7554/eLife.29656
9. Wang W, Racioppi C, Gravez B, Christiaen L. Purification of fluorescent labeled cells from dissociated Ciona embryos. Advances in Experimental Medicine and Biology, 2018;1029: 101–107.
10. Wang W, Zhang Q, Guo D. An Artemisia WD40-repeat gene regulates multiple cellular functions in Arabidopsis. Journal of Biosciences and Medicines (Irvine), 2016;04: 30–36.
11. Razy-Krajka F, Lam K, Wang W, Stolfi A, Joly M, Bonneau R, et al. Collier/OLF/EBF-dependent transcriptional dynamics control pharyngeal muscle specification from primed cardiopharyngeal progenitors. Developmental Cell, 2014;29: 263–276.
12. Wang W, Razy-Krajka F, Siu E, Ketcham A, Christiaen L. NK4 antagonizes Tbx1/10 to promote cardiac versus pharyngeal muscle fate in the ascidian second heart field. PLOS Biology, 2013;11: e1001725.
13. Wang W, Christiaen L. Transcriptional enhancers in ascidian development. Current Topics in Developmental Biology, 2012;98: 147–172.
14. Wu W, Yuan M, Zhang Q, Zhu Y, Yong L, Wang W, et al. Chemotype-dependent metabolic response to methyl jasmonate elicitation inArtemisia annua. Planta Medica, 2011;77: 1048–1053.
15. Wang W, Wang Y, Zhang Q, Qi Y, Guo D. Global characterization of Artemisia annua glandular trichome transcriptome using 454 pyrosequencing. BMC Genomics, 2009;10: 465.[11–15]


 English
English 中文
中文


