New Intron Splicing Code Revealed in Tunicates
A Sars study shows an extraordinary case in which >90% introns have unusual and highly variable splice sites. A new splicing code appeared, which required an evolution of the splicing machinery.
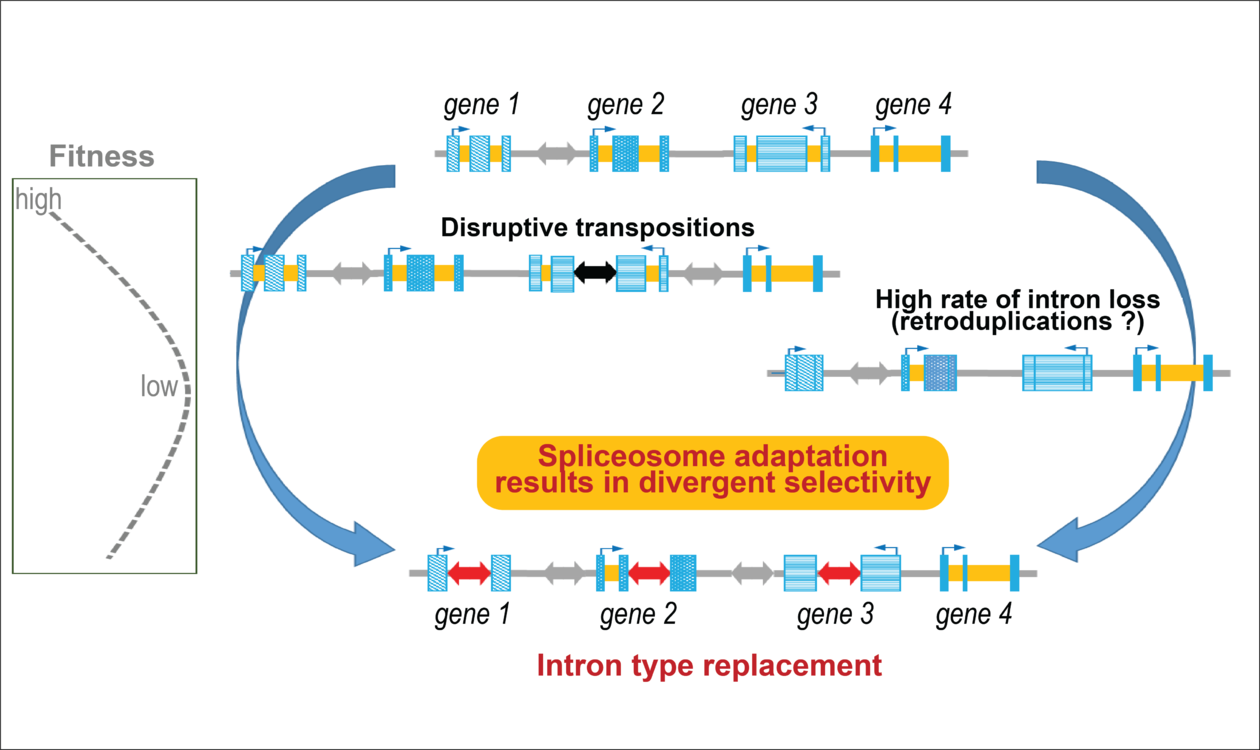
Some tunicate species, including Fritillaria borealis have lost most of their introns, possibly due to highly frequent gene duplications through an RNA mediated mechanism. Transposons can provide introns to compensate the loss, with the risk to inactivate the genes in which they get inserted. A proper evolution of the splicing machinery ensures a correct recognition and removal of the novel transposon-derived introns, which display highly divergent and diverse splice sites.
In principle, almost all introns have canonical sites
Before the RNA copy of a gene can be exported outside the nuclei and translated into a protein, some non-coding segments of the RNA called introns must be precisely recognized and removed while exons are joined together. The process is called RNA splicing and is performed by a complex nuclear machinery called spliceosome, composed of many small RNAs and proteins. For the spliceosome to recognize and process introns, those begin with the dinucleotide GT and end with the dinucleotide AG. In human, almost all introns are GT-AG. A few other introns, called non-canonical, have other types of ends. In human, those are processed by a second and alternative spliceosome. By using various non-standard organisms in its research, the Sars Centre repeatedly found deviations from the main rules. The tunicate Oikopleura dioica, a very important component of the zooplankton in all oceans, shows remarkable genetic properties due to a very rapidly evolving genome. In particular, it was known that many introns of its genes have been newly gained and quite a few are nearly-canonical (GnonT-AG).
A very unexpected deviation from the paradigm
By exploring other species of the same group, an extraordinary situation was found in Fritillaria borealis where non-canonical introns have almost totally replaced the canonical ones. Analyses show that most of these non-canonical introns are mostly derived from small mobile genetic elements (transposons), the insertion of which can destroy active genes. This is not happening here, because the splicing machinery is able to recognize and excise them from gene RNA transcripts. The study shows that for doing this, the spliceosome of Fritillaria has evolved specific properties, which in the end make the transposon insertions inocuitous. For reasons that are not entirely clear, most genes need introns for their expression, and in this case a massive loss of old canonical introns could be overcome by recruiting new introns from mobile elements. Overall, this finding illustrates how during evolution, fundamental processes of gene expression can be modified. The future should tell us more about the circumstances that allowed or made this to happen.
Interview with Daniel Chourrout about this and other recent papers:
In what ways does this new article bring significant new knowledge?
We consider the finding of this article important, for two main reasons. It shows a massive replacement of old introns, relatively recent and so massive that sequence analysis can point to the donor elements. These are non-autonomous DNA transposons. That transposons become introns has been shown previously, including by ourselves in another tunicate species. It was however difficult to estimate the real importance of this intron gain mechanism, as only a few tens of introns in the whole genome were for sure derived from mobile elements. Here, there are thousands of them and therefore, we can imagine from this case that many of old introns, including many of ours, may have appeared in such a way.
There is a second side in this paper, which in our opinion, is more original. Most of these new introns came with non-canonical splice sites, very different from the GT-AG that massively prevail in all genomes. GT-AG introns are here in only a small minority. Strikingly, non-canonical introns are not only numerous, they are also very diverse. Splice sites are not conserved even though some (AG-AC or AG-AT) are more frequent than others. The immediate question was how can they be recognized and removed from transcripts? The study was also experimental, and did not stop after sequence analysis. We showed with two methods (lariat-seq, genome wide effect of spliceosome inhibitors) that the spliceosome is involved. Another type of experiment, introduction of these introns into cells in native or mutated forms, shows that human cells cannot splice them out. Therefore the spliceosome in these tunicates has evolved new properties. With genome mining and well targeted sequencing assays, we could not see what changes in small RNAs or proteins gave the spliceosome such exceptional properties.
What guided you to such investigations?
Since we analyzed the Oikopleura genome and showed its considerable divergence with those of other metazoans, we have been interested in the turnover of introns, combining intron losses and intron gains. While it is generally accepted that most intron losses result from one mechanism (genomic integration of reverse copies of transcripts), multiple scenarios have been proposed to explain intron gains. In Oikopleura, we found indications from four pair of introns, that introns may propagate through a mechanism called reverse splicing, thus far known only for other types of introns. During the last years, we intensified our efforts of genome sequencing for species of the same group, collected around the world. We observed that all the species are the outcome of rapid evolution and intron positions also changed a lot in all these lineages. The first look at the Fritillaria borealis genome and its alignment with transcriptomes, immediately revealed the phenomenon on which this article is based.
In a short time, your group has published three papers in Current Biology? are these papers related to each other in some way?
Yes indeed, because they all show a divergence of mechanisms that cause genome changes. Beyond standard genome analysis, we have developed experimental approaches that bring evidence for and a first characterization of these mechanisms. The first paper (Deng et al, 2018) was demonstrating functionally that a universally conserved pathway of DNA repair (NHEJ) has been lost and replaced by an alternative mechanism that is more mutagenic. The second paper (Naville et al, 2019),in collaboration with JN Volff in ENS Lyon, was reporting considerable variations of genome size resulting from a differential accumulation of non-autonomous retroelements. The third and present one is about massive changes of intron types and shows a necessary evolution of the splicing machinery to process non-canonical ones.
Are these stories ending here or do important questions remain?
As often in science, experimental work provide interesting answers but opens many more questions. We revealed the existence of alternative mechanisms, but we do not know exactly what these mechanisms are. The main question is: are they alternative mechanisms that operate at very low frequency in other more “orthodox” organisms, or were they invented by tunicate larvaceans? We have undertaken research to elucidate some of the molecular pathways related to genome changes, but this turns out to be very challenging technically. The main type of question, even more interesting, is about the reasons for such changes. Is it only a predictable effect of rapid evolution or was there some necessity for evolving divergently. For example, how could these animals lose such a fundamental mechanism of DNA repair ? Why did they lose almost all their old introns, resulting in a need for their replacement, and for this use the risky solution of transposons allowed to enter the genes ? For this second type of question, we have some ideas, but it will be very difficult to find firm evidence for a given scenario.
Any special satisfaction coming from this recent genome research?
Yes, there are many. To convince editors that such research on non-standard model organisms can open new ways of thinking is always difficult. Then, reviewers often want to know more but can be frustrated because such organisms are not amenable to many sorts of functional assays. Here, we have taken the challenge of bringing experimental evidence and not just speculations based on genome mining. These papers result from a handful of authors, most from our small group. Simon Henriet who is author of the three papers had a fantastic contribution. Wei Deng, also did very well in his project on DNA repair. Genomes were assembled by Sara Sumic and she and I did most of the genome mining. Small groups with experienced, smart and rigorous people are able to bring highly significant contributions!


 English
English 中文
中文

