Species-resolved sequencing of low-biomass or degraded microbiomes using 2bRAD-M
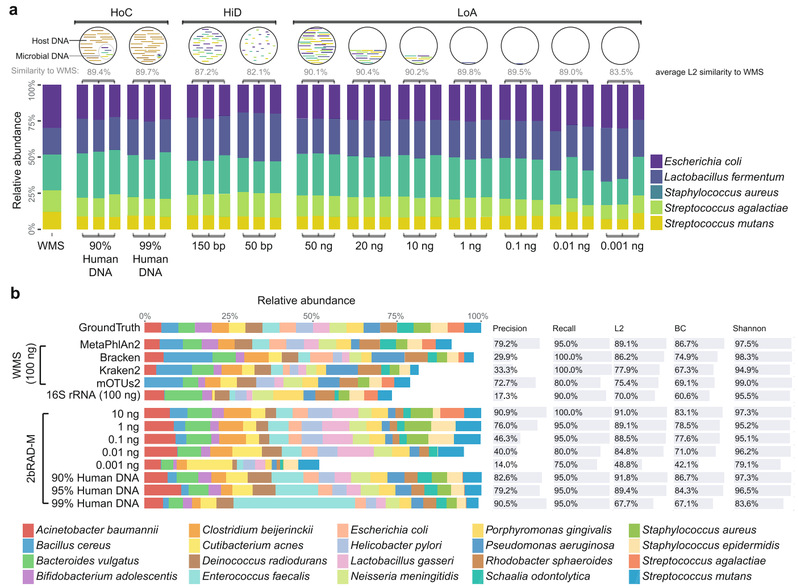
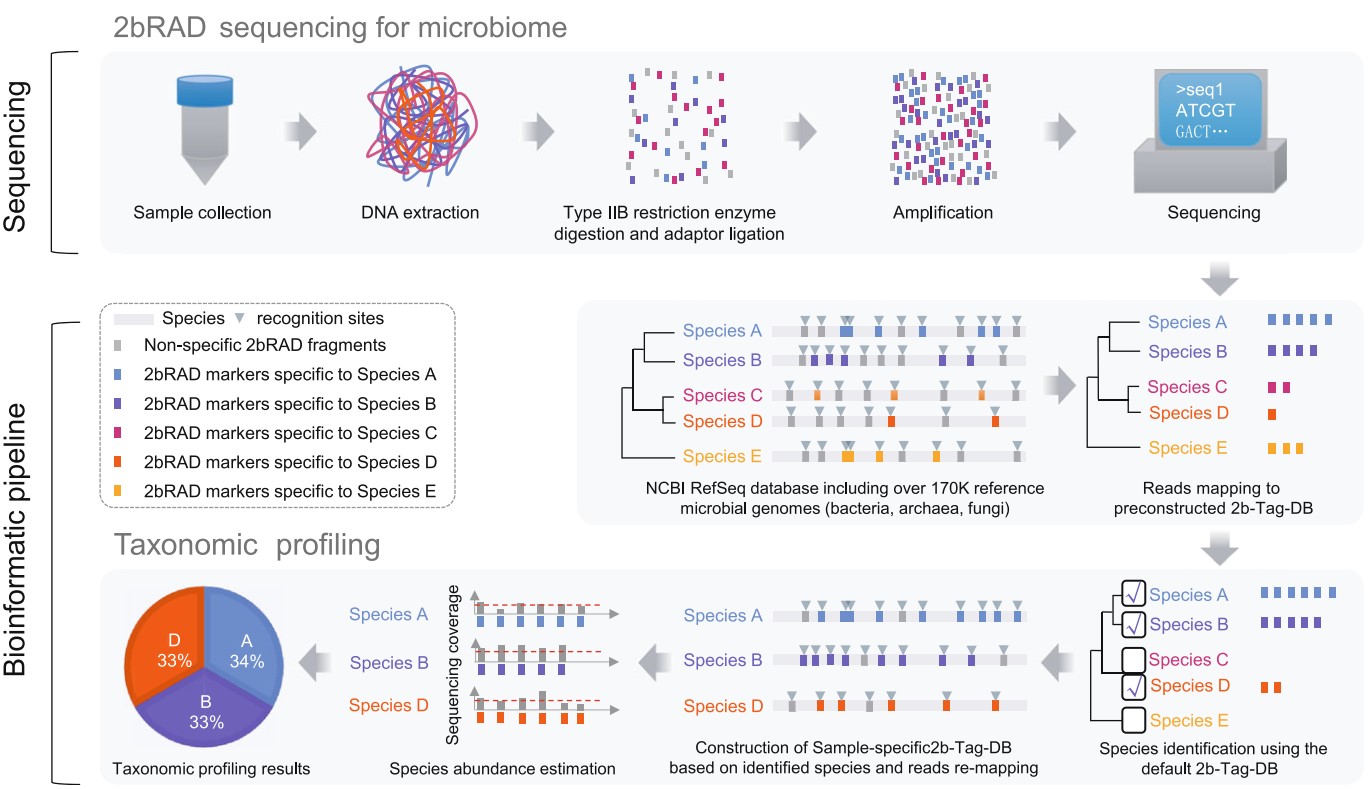
Microbiome samples with low microbial biomass or severe DNA degradation remain challenging for amplicon-based or whole-metagenome sequencing approaches. Here, we introduce 2bRAD-M, a highly reduced and cost-effective strategy which only sequences ~ 1% of metagenome and can simultaneously produce species-level bacterial, archaeal, and fungal profiles. 2bRAD-M can accurately generate species-level taxonomic profiles for otherwise hard-to-sequence samples with merely 1 pg of total DNA, high host DNA contamination, or severely fragmented DNA from degraded samples. Tests of 2bRAD-M on various stool, skin, environmental, and clinical FFPE samples suggest a successful reconstruction of comprehensive, high-resolution microbial profiles. That is what Shi Wang from Sars-Fang Centre, Ocean University of China and Jian Xu from The Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) have published in Genome Biology.


 English
English 中文
中文

